|
Erik Knudsen, Ph.D.: RB and CCND1
Current research is focused on four specific areas
- Cyclin D1 is a proto-oncogene which functions to drive cell cycle progression. The human cyclin D1 gene has a common polymorphism at nucleotide 870 (A/G). The A polymorphism promotes aberrant splicing (producing transcript b), and both the polymorphism and transcript b have been linked to cancer predisposition and poor clinical outcome. We are focusing on modeling the cyclin D1 polymorphism and resultant aberrant splice product in murine models.
As part of our efforts we have cloned the human transcript b. We have confirmed the appropriate transcript by sequencing and are presently characterizing the product for its influence on cell cycle progression, cellular transformation and over-coming the activity of the retinoblastoma tumor suppressor, RB.
Through the consortium we have been collaborating with the MD Anderson group in Smithville Texas. They are currently working on producing mice that express the human transcript b under the control of a keratin promoter. This will enable us to evaluate the specific effect of this allele in a variety of epithelial tissues and to monitor the influence of UV and other environmental stresses upon the polymorphism encoded product.
- RB is a tumor suppressor that is inactivated by multiple mechanisms in the majority of human cancers. Importantly, RB activity is compromised in patients with low-penetrance alleles of RB, wherein either genetic or environmental effects influence whether individuals will develop tumors. The most common low-penetrance tumor mutation of RB is R661W. We have produced the murine equivalent of R661W and are confirming that it compromises RB function using in vitro assays. We have also cloned murine genomic DNA inclusive of exon 20 (the site of the R661 codon) and are in the process of developing targeting constructs to introduce this allele into ES cells.
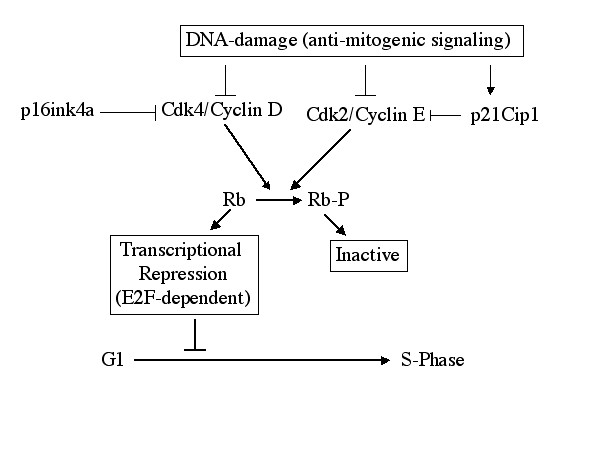
- As noted above, RB is a critical regulator of cell cycle and it is inactivated through a variety of means during tumorigenesis. Additionally, a number of the murine models being developed will likely impact on the RB-pathway (e.g. the cyclin D1, p16ink4a, and RB mice discussed in this center, as well as those being developed in other centers). Biochemically, RB functions as a transcriptional repressor; therefore, to understand how RB functions and to define markers for inactivation in tumor models we carried out a genome-wide analysis of transcriptional targets down-stream of RB. This analysis defined greater than 200 new targets for RB and our lab has determined that many of these targets are down-regulated at the level of protein. Additionally, RB-disruption was shown to lead to the activation of specific targets, which should be particularly useful in the analysis of murine models wherein RB signaling is disrupted. These studies were carried out in concert with Dr. Bruce Aronow who provided the bio-informatics expertise. The data is posted on a web-based platform and should be accessible to all consortium members. Additionally, these studies have been submitted for publication in the journal Cancer Cell.
- The p16ink4a tumor suppressor is also lost at high-frequency in human cancers and functions in concert with cyclin D1 and RB to regulate cell cycle progression. Only this year has it become clear that p16ink4a functions as a tumor suppressor in mice. We had proposed to produce mice harboring alleles of p16ink4a which pre-dispose to melanoma in humans. Now that the null allele of p16ink4a has a defined phenotype, we are gathering the reagents to produce mice with differing alleles of p16ink4a.
Interim Progress
Cyclin D1:
To determine how the cyclin D1 A/G870 polymorphism contributes to tumorigenesis in humans, we initially cloned the cyclin D1 transcript B cDNA that is associated with the A/A genotype. Using the cloned transcript B we have shown that this variant of cyclin D1 has the canonical activities associated with cyclin D1 and additionally harbors the ability to transform 3T3 cells. Interestingly, the transformation phenotype is associated with slower proliferation rates. This mechanistic dissection is about to be submitted for publication. Due to a lack of conservation between the human and murine cyclin D1 transcript B, we are taking a transgenic approach to investigate the function of the polymorphism. In collaboration with MD Anderson Science Park, we have made Keratin 5-Cyclin D1b transgenic mice and my own laboratory is making the MMTV-Cyclin D1b transgenic mice.
RB:
As an initial step in analyzing models with RB compromised in animals we undertook a genome-wide analysis for repression targets of RB. These studies were recently accepted for publication in Cancer Research, and the data is available for all members of the consortium. In terms of generating the animal models, we have cloned 8.0 Kb of genomic RB (in collaboration with Dr. Sanchez’s group) and are in the process of introducing the point mutations for the targeting construct. In all likelihood the construct will be finished by the end of this year.
|
|